-Search query
-Search result
Showing 1 - 50 of 582 items for (author: lim & k)
EMDB-42478: 
Trehalose Synthase (TreS) of Mycobacterium tuberculosis in complex with 6-TreAz compound
Method: single particle / : Pathirage R, Ronning DR

PDB-8uqv: 
Trehalose Synthase (TreS) of Mycobacterium tuberculosis in complex with 6-TreAz compound
Method: single particle / : Pathirage R, Ronning DR
EMDB-19042: 
MAP7 MTBD (microtubule binding domain) decorated microtubule protofilament
Method: single particle / : Bangera M, Moores CA

EMDB-19043: 
MAP7 MTBD (microtubule binding domain) decorated Taxol stabilized microtubule (13pf) protofilament
Method: single particle / : Bangera M, Moores CA

EMDB-19044: 
MAP7 MTBD (microtubule binding domain) decorated Taxol stabilized microtubule (14pf) protofilament
Method: single particle / : Bangera M, Moores CA

PDB-8rc1: 
MAP7 MTBD (microtubule binding domain) decorated microtubule protofilament
Method: single particle / : Bangera M, Moores CA

EMDB-40589: 
hPAD4 bound to Activating Fab hA362
Method: single particle / : Maker A, Verba KA

EMDB-40590: 
hPAD4 bound to inhibitory Fab hI365
Method: single particle / : Maker A, Verba KA

EMDB-34314: 
SARS-CoV-2 RNA E-RTC complex with RMP-nsp9 and GMPPNP
Method: single particle / : Yan LM, Huang YC, Ge J, Liu ZY, Gao Y, Rao ZH, Lou ZY

EMDB-34316: 
SARS-CoV-2 E-RTC bound with MMP-nsp9 and GMPPNP
Method: single particle / : Yan LM, Rao ZH, Lou ZY

PDB-8gwk: 
SARS-CoV-2 RNA E-RTC complex with RMP-nsp9 and GMPPNP
Method: single particle / : Yan LM, Huang YC, Ge J, Liu ZY, Gao Y, Rao ZH, Lou ZY

PDB-8gwm: 
SARS-CoV-2 E-RTC bound with MMP-nsp9 and GMPPNP
Method: single particle / : Yan LM, Rao ZH, Lou ZY

EMDB-19687: 
GroEL with bound GroTAC peptide
Method: single particle / : Wroblewski K, Izert-Nowakowska MA, Goral TK, Klimecka MM, Kmiecik S, Gorna MW

PDB-8s32: 
GroEL with bound GroTAC peptide
Method: single particle / : Wroblewski K, Izert-Nowakowska MA, Goral TK, Klimecka MM, Kmiecik S, Gorna MW
EMDB-42187: 
Eastern equine encephalitis virus (PE-6) VLP (icosahedral)
Method: single particle / : Adams LJ, Fremont DH

EMDB-42189: 
Eastern equine encephalitis virus (PE-6) VLP in complex with full-length VLDLR (icosahedral)
Method: single particle / : Adams LJ, Fremont DH
EMDB-42191: 
Eastern equine encephalitis virus (PE-6) VLP in complex with VLDLR LA(1-2) (icosahedral)
Method: single particle / : Adams LJ, Fremont DH

EMDB-42193: 
Eastern equine encephalitis virus (PE-6) VLP in complex with VLDLR LA(1-3) (icosahedral)
Method: single particle / : Adams LJ, Fremont DH
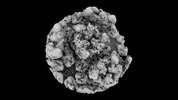
EMDB-42194: 
Eastern equine encephalitis virus (PE-6) VLP in complex with VLDLR LA(1-3) (asymmetric unit)
Method: single particle / : Adams LJ, Fremont DH

EMDB-42195: 
Eastern equine encephalitis virus (PE-6) VLP in complex with VLDLR LA(1-5)[W132A] (icosahedral)
Method: single particle / : Adams LJ, Fremont DH

EMDB-42196: 
Eastern equine encephalitis virus (PE-6) VLP in complex with VLDLR LA(1-5)[W132A] (asymmetric unit)
Method: single particle / : Adams LJ, Fremont DH
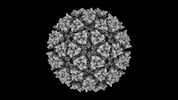
EMDB-42197: 
Eastern equine encephalitis virus (PE-6) VLP in complex with VLDLR LA(1-6)[W132A, W210A] (icosahedral)
Method: single particle / : Adams LJ, Fremont DH

EMDB-42198: 
Eastern equine encephalitis virus (PE-6) VLP in complex with VLDLR LA(1-6)[W132A, W210A] (asymmetric unit)
Method: single particle / : Adams LJ, Fremont DH
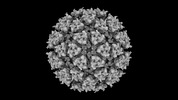
EMDB-42199: 
Eastern equine encephalitis virus (PE-6) VLP in complex with VLDLR LA(1-8)[W132A, W210A, W256A] (icosahedral)
Method: single particle / : Adams LJ, Fremont DH

EMDB-42200: 
Eastern equine encephalitis virus (PE-6) VLP in complex with VLDLR LA(1-8)[W132A, W210A, W256A] (asymmetric unit)
Method: single particle / : Adams LJ, Fremont DH
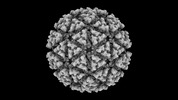
EMDB-42201: 
Eastern equine encephalitis virus (PE-6) VLP in complex with VLDLR LA(3-8) (icosahedral)
Method: single particle / : Adams LJ, Fremont DH

EMDB-42202: 
Eastern equine encephalitis virus (PE-6) VLP in complex with VLDLR LA(1-6)[W89A] (icosahedral)
Method: single particle / : Adams LJ, Fremont DH

EMDB-42203: 
Eastern equine encephalitis virus (PE-6) VLP in complex with VLDLR LA(1-6)[W89A] (asymmetric unit)
Method: single particle / : Adams LJ, Fremont DH
EMDB-42212: 
Eastern equine encephalitis virus (FL93-939) VLP in complex with VLDLR LA(1-6) (icosahedral)
Method: single particle / : Adams LJ, Fremont DH

EMDB-35122: 
Human TRiC-PhLP2A complex in the closed state
Method: single particle / : Roh SH, Park J, Kim H, Lim S

PDB-8i1u: 
Human TRiC-PhLP2A complex in the closed state
Method: single particle / : Roh SH, Park J, Kim H, Lim S

EMDB-35199: 
The focused refinement of CCT3-PhLP2A from TRiC-PhLP2A complex in the open state
Method: single particle / : Roh SH, Park J, Kim H, Lim S

PDB-8i6j: 
The focused refinement of CCT3-PhLP2A from TRiC-PhLP2A complex in the open state
Method: single particle / : Roh SH, Park J, Kim H, Lim S

EMDB-37156: 
Graphene sandwiched equine spleen apoferritin
Method: single particle / : Liu N, Xu J, Wang HW

EMDB-37158: 
Graphene sandwiched 20S proteasome
Method: single particle / : Liu N, Xu J, Wang HW

EMDB-38213: 
Graphene sandwiched spike
Method: single particle / : Liu N, Xu J, Wang HW

EMDB-42188: 
Eastern equine encephalitis virus (PE-6) VLP (asymmetric unit)
Method: single particle / : Adams LJ, Fremont DH

EMDB-42190: 
Eastern equine encephalitis virus (PE-6) VLP in complex with full-length VLDLR (asymmetric unit)
Method: single particle / : Adams LJ, Fremont DH

EMDB-42192: 
Eastern equine encephalitis virus (PE-6) VLP in complex with VLDLR LA(1-2) (asymmetric unit)
Method: single particle / : Adams LJ, Fremont DH

PDB-8ufa: 
Eastern equine encephalitis virus (PE-6) VLP (asymmetric unit)
Method: single particle / : Adams LJ, Fremont DH, Center for Structural Biology of Infectious Diseases (CSBID)

PDB-8ufb: 
Eastern equine encephalitis virus (PE-6) VLP in complex with full-length VLDLR (asymmetric unit)
Method: single particle / : Adams LJ, Fremont DH, Center for Structural Biology of Infectious Diseases (CSBID)

PDB-8ufc: 
Eastern equine encephalitis virus (PE-6) VLP in complex with VLDLR LA(1-2) (asymmetric unit)
Method: single particle / : Adams LJ, Fremont DH, Center for Structural Biology of Infectious Diseases (CSBID)

EMDB-35284: 
Human TRiC-PhLP2A complex in the open state
Method: single particle / : Roh SH, Park J, Kim H, Lim S

PDB-8i9u: 
Human TRiC-PhLP2A complex in the open state
Method: single particle / : Roh SH, Park J, Kim H, Lim S

EMDB-29296: 
Cryo-EM structure of NLR family apoptosis inhibitory protein 5 (NAIP5) in complex with a full-length flagellin (FliC) ligand
Method: single particle / : Paidimuddala B, Zhang L

PDB-8fml: 
Cryo-EM structure of NLR family apoptosis inhibitory protein 5 (NAIP5) in complex with a full-length flagellin (FliC) ligand
Method: single particle / : Paidimuddala B, Zhang L

EMDB-16659: 
Inward-facing conformation of the ABC transporter BmrA C436S/A582C cross-linked mutant
Method: single particle / : Hanssen E, Valimehr S, Di Cesare M, Kaplan E, Orelle C, Jault JM

EMDB-18535: 
Inward-facing conformation of the ABC transporter BmrA
Method: single particle / : Hanssen E, Valimehr S, Di Cesare M, Kaplan E, Orelle C, Jault JM
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search





 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
